03 - Quantum Kernel Machine Learning
[1]:
# This code is from:
# https://qiskit-community.github.io/qiskit-machine-learning/tutorials/03_quantum_kernel.html
[2]:
from qiskit_machine_learning.utils import algorithm_globals
algorithm_globals.random_seed = 12345
[3]:
from qiskit_machine_learning.datasets import ad_hoc_data
adhoc_dimension = 2
train_features, train_labels, test_features, test_labels, adhoc_total = ad_hoc_data(
training_size=20,
test_size=5,
n=adhoc_dimension,
gap=0.3,
plot_data=False,
one_hot=False,
include_sample_total=True,
)
[4]:
import matplotlib.pyplot as plt
import numpy as np
def plot_features(ax, features, labels, class_label, marker, face, edge, label):
# A train plot
ax.scatter(
# x coordinate of labels where class is class_label
features[np.where(labels[:] == class_label), 0],
# y coordinate of labels where class is class_label
features[np.where(labels[:] == class_label), 1],
marker=marker,
facecolors=face,
edgecolors=edge,
label=label,
)
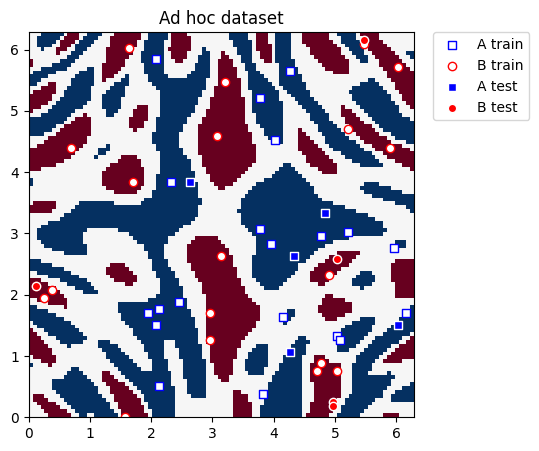
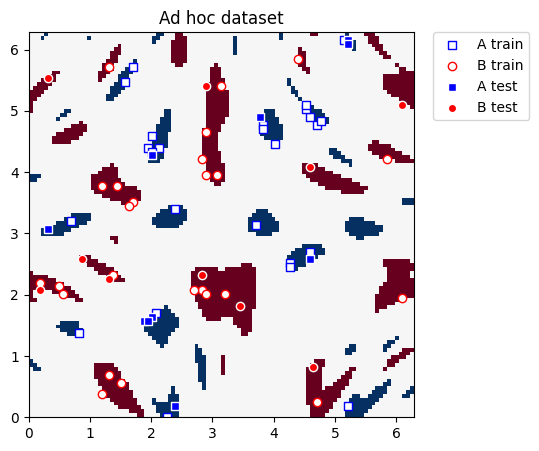
def plot_dataset(train_features, train_labels, test_features, test_labels, adhoc_total):
plt.figure(figsize=(5, 5))
plt.ylim(0, 2 * np.pi)
plt.xlim(0, 2 * np.pi)
plt.imshow(
np.asmatrix(adhoc_total).T,
interpolation="nearest",
origin="lower",
cmap="RdBu",
extent=[0, 2 * np.pi, 0, 2 * np.pi],
)
# A train plot
plot_features(plt, train_features, train_labels, 0, "s", "w", "b", "A train")
# B train plot
plot_features(plt, train_features, train_labels, 1, "o", "w", "r", "B train")
# A test plot
plot_features(plt, test_features, test_labels, 0, "s", "b", "w", "A test")
# B test plot
plot_features(plt, test_features, test_labels, 1, "o", "r", "w", "B test")
plt.legend(bbox_to_anchor=(1.05, 1), loc="upper left", borderaxespad=0.0)
plt.title("Ad hoc dataset")
plt.show()
[5]:
plot_dataset(train_features, train_labels, test_features, test_labels, adhoc_total)
[6]:
from qiskit.circuit.library import ZZFeatureMap
#from qiskit.primitives import Sampler
from qiskit_machine_learning.state_fidelities import ComputeUncompute
#from qiskit_machine_learning.kernels import FidelityQuantumKernel
from quantumrings.toolkit.qiskit import QrSamplerV1 as Sampler
from quantumrings.toolkit.qiskit.machine_learning import QrFidelityQuantumKernel
adhoc_feature_map = ZZFeatureMap(feature_dimension=adhoc_dimension, reps=2, entanglement="linear")
sampler = Sampler()
fidelity = ComputeUncompute(sampler=sampler)
#adhoc_kernel = FidelityQuantumKernel(fidelity=fidelity, feature_map=adhoc_feature_map)
adhoc_kernel = QrFidelityQuantumKernel(fidelity=fidelity, feature_map=adhoc_feature_map)
C:\Users\vkasi\AppData\Local\Temp\ipykernel_12608\2195999342.py:14: DeprecationWarning: V1 Primitives are deprecated as of qiskit-machine-learning 0.8.0 and will be removed no sooner than 4 months after the release date. Use V2 primitives for continued compatibility and support.
fidelity = ComputeUncompute(sampler=sampler)
[7]:
from sklearn.svm import SVC
adhoc_svc = SVC(kernel=adhoc_kernel.evaluate)
adhoc_svc.fit(train_features, train_labels)
adhoc_score_callable_function = adhoc_svc.score(test_features, test_labels)
print(f"Callable kernel classification test score: {adhoc_score_callable_function}")
Callable kernel classification test score: 1.0
[8]:
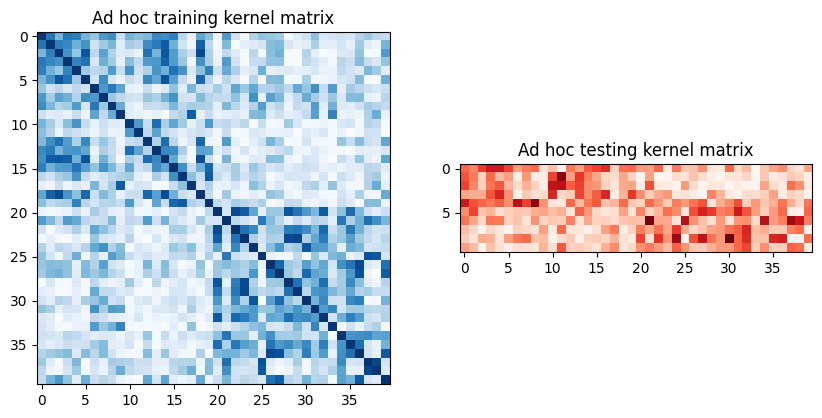
adhoc_matrix_train = adhoc_kernel.evaluate(x_vec=train_features)
adhoc_matrix_test = adhoc_kernel.evaluate(x_vec=test_features, y_vec=train_features)
fig, axs = plt.subplots(1, 2, figsize=(10, 5))
axs[0].imshow(
np.asmatrix(adhoc_matrix_train), interpolation="nearest", origin="upper", cmap="Blues"
)
axs[0].set_title("Ad hoc training kernel matrix")
axs[1].imshow(np.asmatrix(adhoc_matrix_test), interpolation="nearest", origin="upper", cmap="Reds")
axs[1].set_title("Ad hoc testing kernel matrix")
plt.show()
[9]:
adhoc_svc = SVC(kernel="precomputed")
adhoc_svc.fit(adhoc_matrix_train, train_labels)
adhoc_score_precomputed_kernel = adhoc_svc.score(adhoc_matrix_test, test_labels)
print(f"Precomputed kernel classification test score: {adhoc_score_precomputed_kernel}")
Precomputed kernel classification test score: 1.0
[10]:
from qiskit_machine_learning.algorithms import QSVC
qsvc = QSVC(quantum_kernel=adhoc_kernel)
qsvc.fit(train_features, train_labels)
qsvc_score = qsvc.score(test_features, test_labels)
print(f"QSVC classification test score: {qsvc_score}")
QSVC classification test score: 1.0
[11]:
print(f"Classification Model | Accuracy Score")
print(f"---------------------------------------------------------")
print(f"SVC using kernel as a callable function | {adhoc_score_callable_function:10.2f}")
print(f"SVC using precomputed kernel matrix | {adhoc_score_precomputed_kernel:10.2f}")
print(f"QSVC | {qsvc_score:10.2f}")
Classification Model | Accuracy Score
---------------------------------------------------------
SVC using kernel as a callable function | 1.00
SVC using precomputed kernel matrix | 1.00
QSVC | 1.00
[12]:
adhoc_dimension = 2
train_features, train_labels, test_features, test_labels, adhoc_total = ad_hoc_data(
training_size=25,
test_size=0,
n=adhoc_dimension,
gap=0.6,
plot_data=False,
one_hot=False,
include_sample_total=True,
)
[13]:
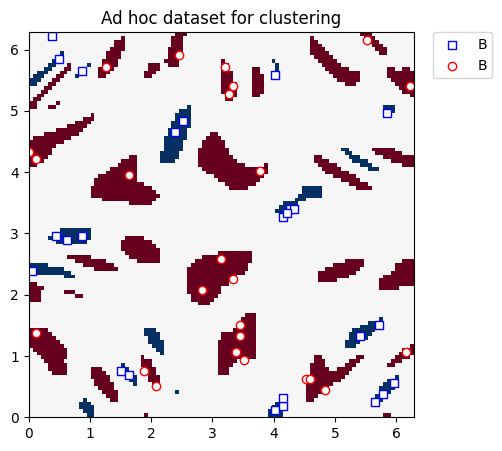
plt.figure(figsize=(5, 5))
plt.ylim(0, 2 * np.pi)
plt.xlim(0, 2 * np.pi)
plt.imshow(
np.asmatrix(adhoc_total).T,
interpolation="nearest",
origin="lower",
cmap="RdBu",
extent=[0, 2 * np.pi, 0, 2 * np.pi],
)
# A label plot
plot_features(plt, train_features, train_labels, 0, "s", "w", "b", "B")
# B label plot
plot_features(plt, train_features, train_labels, 1, "o", "w", "r", "B")
plt.legend(bbox_to_anchor=(1.05, 1), loc="upper left", borderaxespad=0.0)
plt.title("Ad hoc dataset for clustering")
plt.show()
[14]:
adhoc_feature_map = ZZFeatureMap(feature_dimension=adhoc_dimension, reps=2, entanglement="linear")
#adhoc_kernel = FidelityQuantumKernel(feature_map=adhoc_feature_map)
adhoc_kernel = QrFidelityQuantumKernel(feature_map=adhoc_feature_map)
[15]:
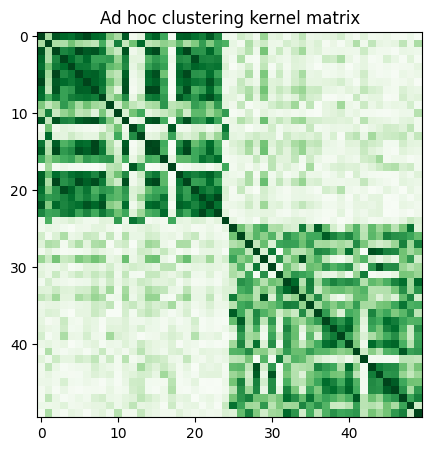
adhoc_matrix = adhoc_kernel.evaluate(x_vec=train_features)
plt.figure(figsize=(5, 5))
plt.imshow(np.asmatrix(adhoc_matrix), interpolation="nearest", origin="upper", cmap="Greens")
plt.title("Ad hoc clustering kernel matrix")
plt.show()
[16]:
from sklearn.cluster import SpectralClustering
from sklearn.metrics import normalized_mutual_info_score
adhoc_spectral = SpectralClustering(2, affinity="precomputed")
cluster_labels = adhoc_spectral.fit_predict(adhoc_matrix)
cluster_score = normalized_mutual_info_score(cluster_labels, train_labels)
print(f"Clustering score: {cluster_score}")
Clustering score: 0.7287008798015754
[17]:
adhoc_dimension = 2
train_features, train_labels, test_features, test_labels, adhoc_total = ad_hoc_data(
training_size=25,
test_size=10,
n=adhoc_dimension,
gap=0.6,
plot_data=False,
one_hot=False,
include_sample_total=True,
)
[18]:
plot_dataset(train_features, train_labels, test_features, test_labels, adhoc_total)
[19]:
feature_map = ZZFeatureMap(feature_dimension=2, reps=2, entanglement="linear")
#qpca_kernel = FidelityQuantumKernel(fidelity=fidelity, feature_map=feature_map)
qpca_kernel = QrFidelityQuantumKernel(fidelity=fidelity, feature_map=feature_map)
[20]:
matrix_train = qpca_kernel.evaluate(x_vec=train_features)
matrix_test = qpca_kernel.evaluate(x_vec=test_features, y_vec=train_features)
[21]:
from sklearn.decomposition import KernelPCA
kernel_pca_rbf = KernelPCA(n_components=2, kernel="rbf")
kernel_pca_rbf.fit(train_features)
train_features_rbf = kernel_pca_rbf.transform(train_features)
test_features_rbf = kernel_pca_rbf.transform(test_features)
kernel_pca_q = KernelPCA(n_components=2, kernel="precomputed")
train_features_q = kernel_pca_q.fit_transform(matrix_train)
test_features_q = kernel_pca_q.transform(matrix_test)
[22]:
from sklearn.linear_model import LogisticRegression
logistic_regression = LogisticRegression()
logistic_regression.fit(train_features_q, train_labels)
logistic_score = logistic_regression.score(test_features_q, test_labels)
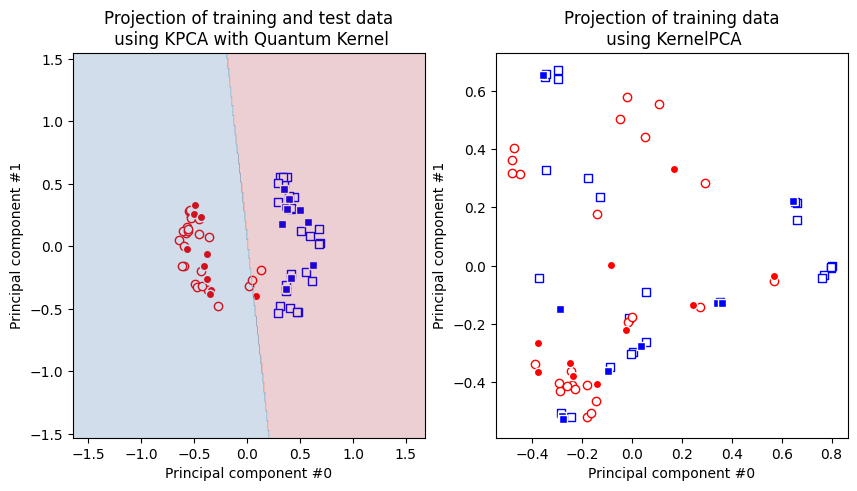
print(f"Logistic regression score: {logistic_score}")
Logistic regression score: 0.95
[23]:
fig, (q_ax, rbf_ax) = plt.subplots(1, 2, figsize=(10, 5))
plot_features(q_ax, train_features_q, train_labels, 0, "s", "w", "b", "A train")
plot_features(q_ax, train_features_q, train_labels, 1, "o", "w", "r", "B train")
plot_features(q_ax, test_features_q, test_labels, 0, "s", "b", "w", "A test")
plot_features(q_ax, test_features_q, test_labels, 1, "o", "r", "w", "A test")
q_ax.set_ylabel("Principal component #1")
q_ax.set_xlabel("Principal component #0")
q_ax.set_title("Projection of training and test data\n using KPCA with Quantum Kernel")
# Plotting the linear separation
h = 0.01 # step size in the mesh
# create a mesh to plot in
x_min, x_max = train_features_q[:, 0].min() - 1, train_features_q[:, 0].max() + 1
y_min, y_max = train_features_q[:, 1].min() - 1, train_features_q[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, h), np.arange(y_min, y_max, h))
predictions = logistic_regression.predict(np.c_[xx.ravel(), yy.ravel()])
# Put the result into a color plot
predictions = predictions.reshape(xx.shape)
q_ax.contourf(xx, yy, predictions, cmap=plt.cm.RdBu, alpha=0.2)
plot_features(rbf_ax, train_features_rbf, train_labels, 0, "s", "w", "b", "A train")
plot_features(rbf_ax, train_features_rbf, train_labels, 1, "o", "w", "r", "B train")
plot_features(rbf_ax, test_features_rbf, test_labels, 0, "s", "b", "w", "A test")
plot_features(rbf_ax, test_features_rbf, test_labels, 1, "o", "r", "w", "A test")
rbf_ax.set_ylabel("Principal component #1")
rbf_ax.set_xlabel("Principal component #0")
rbf_ax.set_title("Projection of training data\n using KernelPCA")
plt.show()
[ ]: